Optimising Essential Particle Dynamics Codes for Multi-Scale Engineering Flow Simulation
eCSE02-02Key Personnel
PI/Co-I: Prof Jason M Reese - University of Edinburgh
Technical: Mr Saif Mulla - University of Edinburgh
Project summary
The primary objective of this project was to optimise essential particle dynamics codes on ARCHER in order to enable researchers to simulate large multi-scale flow problems that had previously been practically unachievable with the existing parallel strategy in our multi-scale software, implemented in OpenFOAM.
Multi-scale technologies have many important practical applications in energy, transportation and healthcare, including: nano-filtering of seawater to make it drinkable for water-stressed populations; re-engineering of aeroplane surfaces to incorporate active micro devices to reduce drag; micro/nano-structuring of ship hull surfaces for breakthrough performance.
Project description
The aims of this project all centred on further development of mdFoam, a software application built upon the Computational Fluid Dynamics (CFD) framework OpenFOAM. mdFoam uses OpenFOAM to run molecular dynamics (MD) simulations of nanoscale and multi-scale flow problems.
mdFoam was developed in the group of the PI, and has been released by CFD Direct Ltd (the OpenFOAM project architects) along with the main OpenFOAM code since 2009. It therefore has a relatively long history and has generated a good track record of research output. Before this project, however, it was under development by a number of different people, each working independently of each other with different aims. The main goal of this project was to improve the parallel scalability of mdFoam by improving one of its major bottlenecks - MPI latency - by hybridising its existing parallelisation strategy with OpenMP per-node parallelisation on the ARCHER platform. Other aims were to start to formalise the software project for release to a wider audience as well as to improve general performance on ARCHER to enable larger-scale simulations, thereby increasing the software's potential for scientific impact.
Although unforeseen circumstances meant that we were not able to produce a new hybrid MPI/OpenMP design within the lifetime of this project, a number of notable successes were achieved. The first is that the key design problems inherent to the underlying OpenFOAM libraries upon which mdFoam is built have been uncovered, and could therefore be addressed in future work. While some of the HPC-related problems that exist within OpenFOAM were known before this project, others were not and have been uncovered as a direct result of the project work. The most important of these is that the Lagrangian OpenFOAM libraries, which mdFoam (and a number of other important codes) require, are not designed with high performance computing in mind, especially the shared memory paradigm. This is due largely to the way in which the libraries lay out memory and treat each unique component of a Lagrangian simulation as a self-contained C++ object, leading to a lack of memory contiguity and severe problems with cache coherency. This problem gives a clear direction for future efforts to speed up not only mdFoam but any other code based on OpenFOAM's Lagrangian code-base.
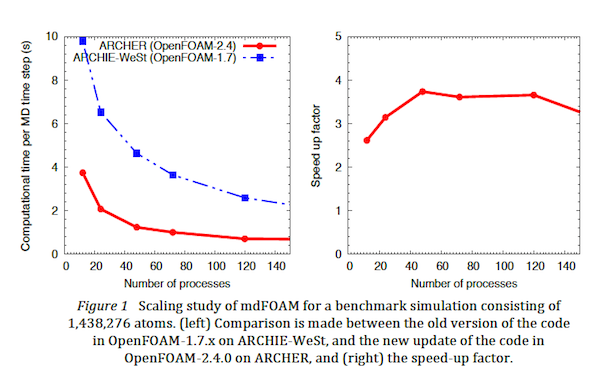
Other important outcomes of this project are that the mdFoam application was upgraded to use a more recent version of the OpenFOAM libraries. This was partly out of necessity of making the code run with the compilers provided by ARCHER, and resulted in a notable 4x speed-up in performance of the code when compared to prior runs using the old code-base on the ARCHIE-WeSt regional Tier-2 system. This can be attributed to general improvements within the OpenFOAM libraries but also a re-working of the cell-linked list lookup implementation in the code, which has changed to better fit the latest Lagrangian OpenFOAM libraries.
Another key outcome is that, following this project, mdFoam has now been released as part of a publicly available GitHub repository (http://www.github.com/MicroNanoFlows), along with other codes produced by the research partnership of the PI and his colleagues, the Micro & Nano Flows for Engineering group, http://www.micronanoflows.ac.uk/. This also includes a number of documented tutorial cases as well as detailed documentation that includes installation instructions for ARCHER, amongst other platforms. The choice of releasing the software via GitHub as a Git repository was as a direct result of this project and can be considered a milestone in the software's history. This will undoubtedly improve the software's potential scientific impact, making it available to researchers outside of its developing research group and facilitating its uptake, as well as contributing to its future sustainability.
Achievement of objectives
- Enabling simulations on ARCHER: Our OpenFOAM code-base, originally forked from the 1.7.x version of OpenFOAM (released in 2012), was not able to run on ARCHER due to compiler obsolescence. The first objective of this project has been met by advancing our entire software base to a more recent version of OpenFOAM (2.4.0), including the molecular dynamics mdFoam code, the direct simulation Monte Carlo (dsmcFoam) code, and the multi-scale software.
- Improved parallel performance: The revision update to the code base involved a revisit of the cell-list algorithm, which is the backbone and consists of the highest computational cost of a molecular dynamics simulation, to deal with better parallelism. As a result we were able to obtain a 4x speed-up of the code on ARCHER (relative to benchmarks on the Tier 2 ARCHIE-WeSt supercomputer), as confirmed by scaling studies over several hundreds of processors, shown in Figure 1.
- Performance problems observed in OpenFOAM: Our scaling experiments revealed some serious bottlenecks in the base OpenFOAM software, including: the large number of individual processor files written to disk, and the badly-written Lagrangian track-to-face function that handles the motion of particles within the domain. A memory contiguity problem in the Cloud class, from which the MD code is derived, has been identified as the main cause behind the lack of success in implementing a hybrid OpenMP-MPI parallelism strategy.
- Sustainable and future-proofing our software: This project has enabled the latest OpenFOAM multi-scale software base to be released to the public on the GitHub website (http://www.github.com/MicroNanoFlows), to be widely used (or further improved) for the benefit of the computational engineering research community.
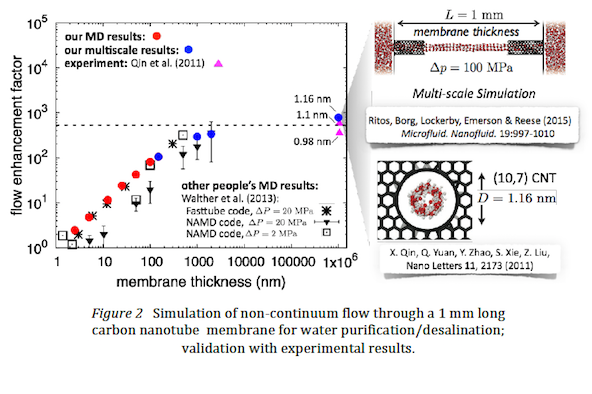
- Scientific insight and impact using multi-scale software: The increased performance on ARCHER for the new code has already enabled us to publish a significant number of high-impact journal papers. One example is the first ever experiment-scale simulation of water flowing through aligned carbon nanotube reverse osmosis membranes [K Ritos, MK Borg, DA Lockerby, DR Emerson & JM Reese (2015) Hybrid molecularcontinuum simulations of water flow through carbon nanotube membranes of realistic thickness, Microfluidics & Nanofluidics, 19:997-1010]. If the same case was run as a full molecular simulation it would take approximately 300 years on a supercomputer to produce the results; using our software and a multi-scale methodology the simulation took 16 weeks. Sample results are shown in Figure 2.
Summary of the software
The software which is produced from this project is available for free to download from a public repository on GitHub (http://www.github.com/MicroNanoFlows). This is intended as a central resource for all the research partnership codes that are included as a part of this repository, including a molecular dynamics code (mdFoam), a direct simulation Monte Carlo code (dsmcFoam), as well as other multi-scale software produced by the partnership, such as the Internal-flow Multiscale Method (IMM). All of these codes are designed and tested to run in parallel on ARCHER. The master branch of the GitHub repository represents the most up-to-date version of our software, including the introduction of a new parallelised cell-list algorithm (incorporated in this project). The repository also includes full documentation of how to use the software, including an integrated Wiki and a 40-page tutorial document along with instructions for installing the software on ARCHER.






